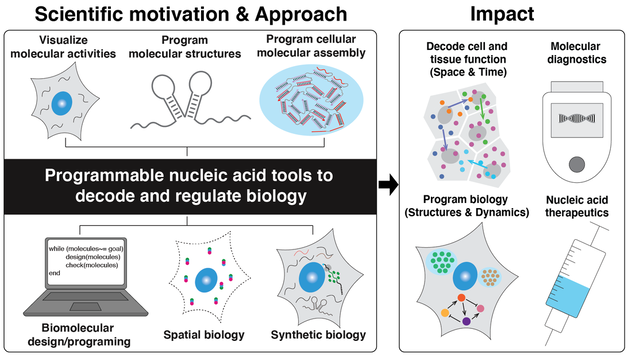
In biological systems, there are billions of biomolecules that containing different levels of biological information, such as primary sequence, folded structural information, function, and chemical connectivity, from single cell to tissue scale. To better understand biological mechanisms and improve human healthcare, it is critical to visualize and manipulate these biomolecular activities in cells. Designing structural and functional biomolecular tools is one of key pillars to achieve these goals. For instance, biomolecular probes created for bio-imaging can help massively visualize biomolecules in cells and tissues, which is necessary for revealing the complex cell behaviors and organizations. Beyond visualization, designed functional biomolecular tools can also regulate cellular activities by interfacing with them to either record cellular information or control cell fates.
Motivated by the scientific challenges to decode and regulate complex biological systems, the research at Hong Lab will move along on the following tracks:
Motivated by the scientific challenges to decode and regulate complex biological systems, the research at Hong Lab will move along on the following tracks:
1. Decoding molecular systems in biology:
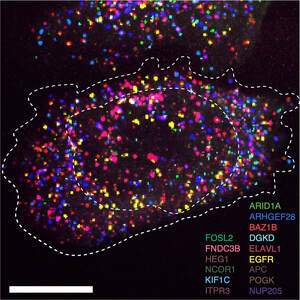
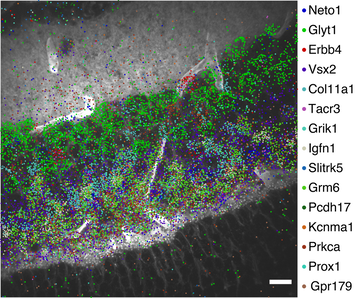
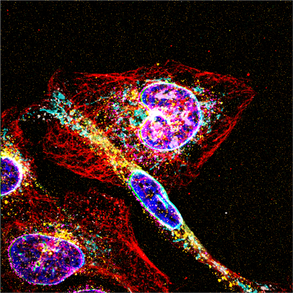
Biological systems are self-organized biomolecules with different species, quantities and spatial locations within cells and tissues. We aim to develop molecular technologies based on programmable DNA reactions on top of fluorescence microscopy and next generation sequencing to analyze biomolecules (e.g., DNAs, RNAs, and proteins) in cells and tissues with high sensitivity, high multiplexity, and high throughput to understand their function.
Biological systems are self-organized biomolecules with different species, quantities and spatial locations within cells and tissues. We aim to develop molecular technologies based on programmable DNA reactions on top of fluorescence microscopy and next generation sequencing to analyze biomolecules (e.g., DNAs, RNAs, and proteins) in cells and tissues with high sensitivity, high multiplexity, and high throughput to understand their function.
References:
1. Fan Hong, Jocelyn Y. Kishi1, Ryan N. Delgado, Jiyoun Jeong, Sinem K. Saka, Hanquan Su, Constance L. Cepko, Peng Yin*. Thermal-plex: Fluidic-free, rapid sequential multiplexed imaging with DNA encoded thermal channels. Nature Methods, 2023, In press.
1. Fan Hong, Jocelyn Y. Kishi1, Ryan N. Delgado, Jiyoun Jeong, Sinem K. Saka, Hanquan Su, Constance L. Cepko, Peng Yin*. Thermal-plex: Fluidic-free, rapid sequential multiplexed imaging with DNA encoded thermal channels. Nature Methods, 2023, In press.
2. Understand/control bio-dynamics
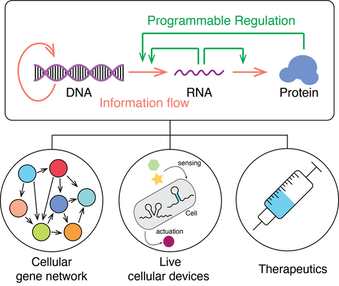
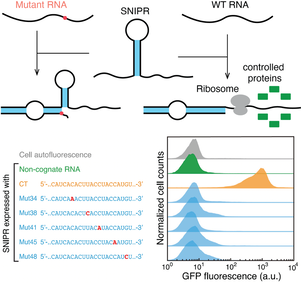
The biological information flow is build upon the central dogma in cells. The regulation of the genome function in nature is through complex biomolecular machinery. One scientific challenge ahead of us is whether can we design those types of biomolecular machinery to control cellular gene expression activities. To address this challenge, we design RNA molecules to hack the central dogma to program cellular gene expression. The regulation mechanism can be used to develop a complex “biomolecular algorithm” that operates in cells, leading to living cellular devices to record the environmental signal, diagnostics, and novel mRNA therapeutic methods.
The biological information flow is build upon the central dogma in cells. The regulation of the genome function in nature is through complex biomolecular machinery. One scientific challenge ahead of us is whether can we design those types of biomolecular machinery to control cellular gene expression activities. To address this challenge, we design RNA molecules to hack the central dogma to program cellular gene expression. The regulation mechanism can be used to develop a complex “biomolecular algorithm” that operates in cells, leading to living cellular devices to record the environmental signal, diagnostics, and novel mRNA therapeutic methods.
References:
- Fan Hong, Petr Sulc. An emergent understanding of strand displacement in RNA biology, Journal of Structural Biology, 2019, 207, 241-249.
- Fan Hong, Duo Ma, Kaiyue Wu, Lida A. Mina, Rebecca C. Luiten, Yan Liu, Hao Yan*, Alexander A. Green*. Precise and Programmable Detection of Mutations Using Ultraspecific Riboregulators, Cell, 2020, 180, 1018-1032.
- Longlong Si, Haiqing Bai, Crystal Yuri Oh, Amanda Jiang, Fan Hong, Tian Zhang, Yongxin Ye, Tristan X Jordan, James Logue, Marisa McGrath, Chaitra Belgur, Karina Calderon, Atiq Nurani, Wuji Cao, Kenneth E Carlson, Rachelle Prantil-Baun, Steven P Gygi, Dong Yang, Colleen B Jonsson, Matthew Frieman, Donald E Ingber, Self-assembling short immunostimulatory duplex RNAs with broad-spectrum antiviral activity, Molecular Therapy-Nucleic Acids, 2022, 29, 923-940.
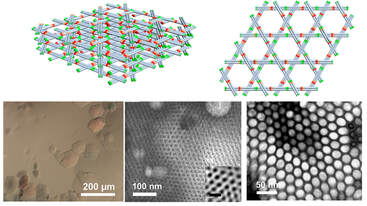
3. Understand/Control bio-matter
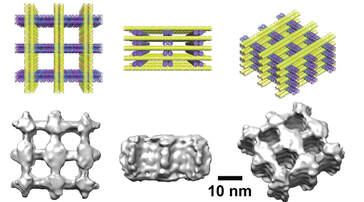
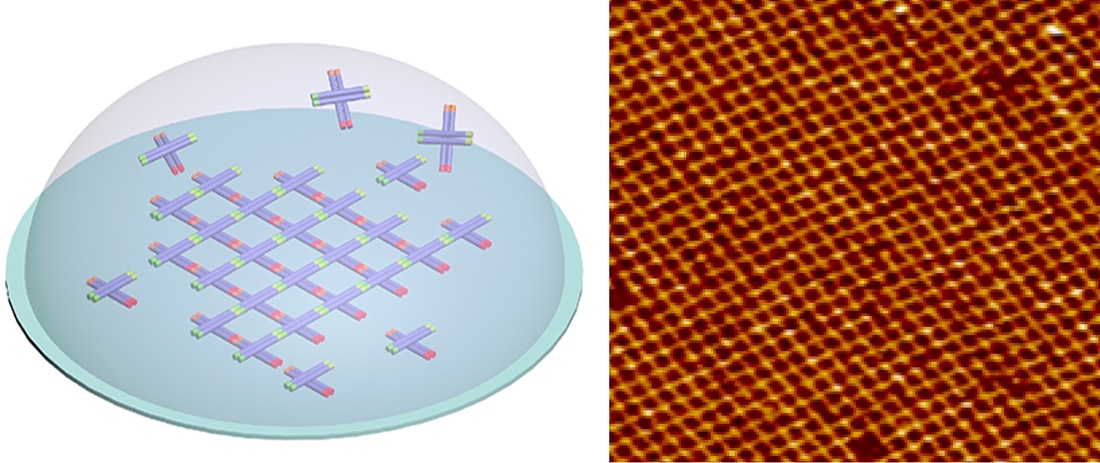
Biomolecules self-organize in cells and the collective self-organization relates to its functions and diseases. We aim to understand and control the collective assembly behavior of nucleic acid both in vivo and in vitro and how it can be use to regulate cell functions. Nucleic acid has highly predictable interactions and can be used as an ideal building blocks to control the molecular assembly in vitro and in vivo. We aim to develop bimolecular tools that allows spatial manipulation within and across the cells to control cellular functions.
Biomolecules self-organize in cells and the collective self-organization relates to its functions and diseases. We aim to understand and control the collective assembly behavior of nucleic acid both in vivo and in vitro and how it can be use to regulate cell functions. Nucleic acid has highly predictable interactions and can be used as an ideal building blocks to control the molecular assembly in vitro and in vivo. We aim to develop bimolecular tools that allows spatial manipulation within and across the cells to control cellular functions.
References:
- Fan Hong, John Shreck, Petr Sulc, Understanding DNA interactions in crowded environments with a coarse-grained model, Nucleic Acid Research, 2020,48,10726.
- Fan Hong, Shuoxing Jiang, Xiang Lan, Raghu Pradeep Narayanan, Petr, Sulc, Fei Zhang, Yan Liu, Hao Yan, Layered-Crossover Tiles with Precisely Tunable Angles for 2D and 3D DNA Crystal Engineering, J. Am. Chem. Soc. 2018, 140, 14670-14676.
- Shuoxing Jiang #, Fan Hong#, Huiyu Hu, Hao Yan, Yan Liu, Understanding the Elementary steps in DNA tile-based self-assembly. ACS nano, 2017, 11,9370-9381. Contributed Equally.
- Fan Hong, Fei Zhang, Yan Liu, Hao Yan,DNA origami: scaffolds for creating high order structures. Chemical Review, 2017, 117, 12584-12640
- Fan Hong, Shuoxing Jiang, Tong Wang, Yan Liu, Hao Yan, 3D Framework DNA origami structure with layered crossover motifs. Angew Chem Int Ed, 2016,55, 12832-12835.